Speed Up your Algorithms Part 2— Numba
Get C++/Fortran like speed for your functions with Numba
This is the third post in a series I am writing. All posts are here:
- Speed Up your Algorithms Part 1 — PyTorch
- Speed Up your Algorithms Part 2 — Numba
- Speed Up your Algorithms Part 3 — Parallelization
- Speed Up your Algorithms Part 4 — Dask
And these goes with Jupyter Notebooks available here:
[Github-SpeedUpYourAlgorithms] and [Kaggle]
Index
- Introduction
- Why Numba?
- How does Numba Works?
- Using basic numba functionalities (Just @jit it!)
- The @vectorize wrapper
- Running your functions on GPU
- Further Reading
- References
NOTE:
This post goes with Jupyter Notebook available in my Repo on Github:[SpeedUpYourAlgorithms-Numba]1. Introduction
Numba is a Just-in-time compiler for python, i.e. whenever you make a call to a python function all or part of your code is converted to machine code “just-in-time” of execution, and it will then run on your native machine code speed! It is sponsored by Anaconda Inc and has been/is supported by many other organisations.
With Numba, you can speed up all of your calculation focused and computationally heavy python functions(eg loops). It also has support for numpy library! So, you can use numpy in your calculations too, and speed up the overall computation as loops in python are very slow. You can also use many of the functions of math library of python standard library like sqrt etc. For a comprehensive list of all compatible functions look here.
2. Why Numba?

[Source]
So, why numba? When there are many other compilers like cython, or any other similar compilers or something like pypy.
For a simple reason that here you don’t have to leave the comfort zone of writing your code in python. Yes, you read it right, you don’t have to change your code at all for basic speedup which is comparable to speedup you get from similar cython code with type definitions. Isn’t that great?
You just have to add a familiar python functionality, a decorator (a wrapper) around your functions. A wrapper for a class is also under development.
So, you just have to add a decorator and you are done. eg:
from numba import jit@jit
def function(x):
# your loop or numerically intensive computations
return x
It still looks like a pure python code, doesn’t it?
3. How does numba work?
Numba generates optimized machine code from pure Python code using LLVM compiler infrastructure. Speed of code run using numba is comparable to that of similar code in C, C++ or Fortran.
Here is how the code is compiled:
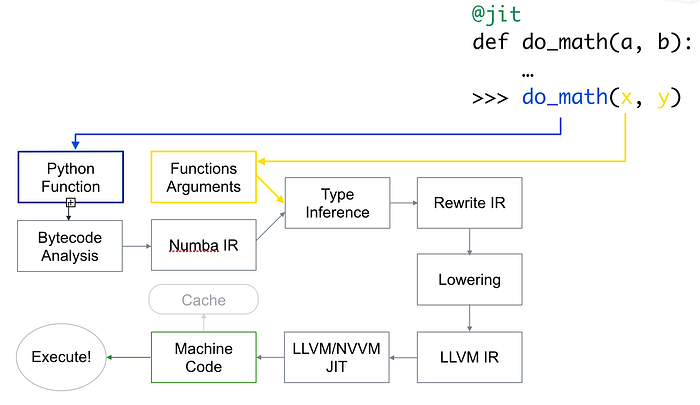
[Source]
First, Python function is taken, optimized and is converted into Numba’s intermediate representation, then after type inference which is like Numpy’s type inference (so python float is a float64), it is converted into LLVM interpretable code. This code is then fed to LLVM’s just-in-time compiler to give out machine code.
You can generate code at runtime or import time on CPU (default) or GPU, as you prefer it.
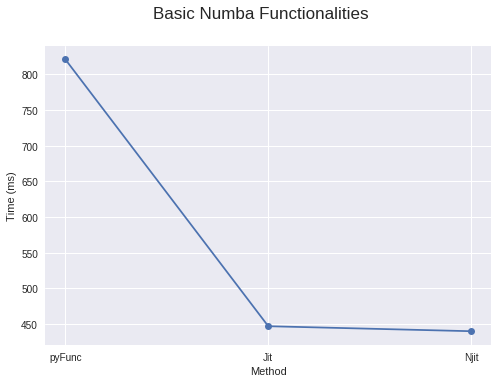
4. Using basic numba functionalities (Just @jit it!)
Piece of cake!
For best performance numba recommends using nopython = True argument with your jit wrapper, using which it won’t use the Python interpreter at all. Or you can also use @njit too. If your wrapper with nopython = True fails with an error, you can use simple @jit wrapper which will compile part of your code, loops it can compile, and turns them into functions, to compile into machine code and give the rest to the python interpreter.
So, you just have to do:
from numba import njit, jit@njit # or @jit(nopython=True)
def function(a, b):
# your loop or numerically intensive computations
return result
When using @jit make sure your code has something numba can compile, like a compute-intensive loop, maybe with libraries (numpy) and functions it supports. Otherwise, it won’t be able to compile anything.
To put a cherry on top, numba also caches the functions after first use as machine code. So after the first time, it will be even faster because it doesn’t need to compile that code again, given that you are using the same argument types that you used before.
And if your code is parallelizable you can also pass parallel = True as an argument, but it must be used in conjunction with nopython = True. For now, it only works on CPU.
You can also specify function signature you want your function to have, but then it won’t compile for any other types of arguments you give to it. For example:
from numba import jit, int32@jit(int32(int32, int32))
def function(a, b):
# your loop or numerically intensive computations
return result# or if you haven't imported type names
# you can pass them as string@jit('int32(int32, int32)')
def function(a, b):
# your loop or numerically intensive computations
return result
Now your function will only take two int32’s and return an int32. By this, you can have more control over your functions. You can even pass multiple functional signatures if you want.
You can also use other wrappers provided by numba:
- @vectorize: allows scalar arguments to be used as numpy
ufuncs, - @guvectorize: produces NumPy generalized
ufuncs, - @stencil: declare a function as a kernel for a stencil-like operation,
- @jitclass: for jit aware classes,
- @cfunc: declare a function for use as a native call back (to be called from C/C++ etc),
- @overload: register your own implementation of a function for use in nopython mode, e.g.
@overload(scipy.special.j0).
Numba also has Ahead of time (AOT) compilation, which produces a compiled extension module which does not depend on Numba. But:
- It allows only regular functions (not ufuncs),
- You have to specify a function signature. You can only specify one, for many specify under different names.
It also produces generic code for your CPU’s architectural family.
5. The @vectorize wrapper
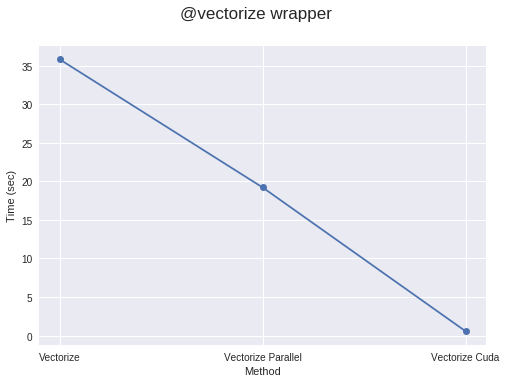
By using @vectorize wrapper you can convert your functions which operate on scalars only, for example, if you are using python’s math library which only works on scalars, to work for arrays. This gives speed similar to that of a numpy array operations (ufuncs). For example:
@vectorize
def func(a, b):
# Some operation on scalars
return resultYou can also pass target argument to this wrapper which can have a value equal to parallel for parallelizing code, cuda for running code on cuda/GPU.
@vectorize(target="parallel")
def func(a, b):
# Some operation on scalars
return resultVectorizing with target = “parallel” or “cuda” will generally run faster than numpy implementation, given your code is sufficiently compute-intensive or array is sufficiently large. If not then it comes with an overhead of the time for making threads and splitting elements for different threads, which can be larger than actual compute time for the whole process. So, work should be sufficiently heavy to get a speedup.
This great video has an example of speeding up Navier Stokes equation for computational fluid dynamics with Numba:
6. Running your functions on GPU
You can also pass @jit like wrappers to run functions on cuda/GPU also. For that, you will have to import cuda from numba library. But running your code on GPU is not going to be as easy as before. It has some initial computations that need to be done for running function on hundreds or even thousands of threads on GPU. You have to declare and manage a hierarchy of grids, blocks and threads. And it’s not that hard.
To execute a function on GPU, you have to either define something called a kernel function or a device function. Firstly let’s see a kernel function.
Some points to remember about kernel functions:
a) kernels explicitly declare their thread hierarchy when called, i.e. the number of blocks and number of threads per block. You can compile your kernel once, and call it multiple times with different block and grid sizes.
b) kernels cannot return a value. So, either you will have to do changes on original array, or pass another array for storing the result. For computing scalar, you will have to pass a 1 element array.
# Defining a kernel function
from numba import cuda@cuda.jit
def func(a, result):
# Some cuda related computation, then
# your computationally intensive code.
# (Your answer is stored in 'result')
So for launching a kernel, you will have to pass two things:
- Number of threads per block,
- Number of blocks.
For example:
threadsperblock = 32
blockspergrid = (array.size + (threadsperblock - 1)) // threadsperblock
func[blockspergrid, threadsperblock](array)Kernel function in every thread has to know in which thread it is, to know which elements of array it is responsible for. Numba makes it easy to get these positions of elements, just by one call.
@cuda.jit
def func(a, result):
pos = cuda.grid(1) # For 1D array
# x, y = cuda.grid(2) # For 2D array
if pos < a.shape[0]:
result[pos] = a[pos] * (some computation)To save the time which will be wasted in copying numpy array to a specific device and then again storing result in numpy array, Numba provides some functions to declare and send arrays to specific device, like: numba.cuda.device_array, numba.cuda.device_array_like, numba.cuda.to_device, etc. to save time of needless copies to cpu(unless necessary).
On the other hand, a device function can only be invoked from inside a device (by a kernel or another device function). The plus point is, you can return a value from a device function. So, you can use this return value of the function to compute something inside a kernel function or a device function.
from numba import cuda@cuda.jit(device=True)
def device_function(a, b):
return a + b
You should also look into supported functionality of Numba’s cuda library, here.
Numba also has implementations of atomic operations, random number generators, shared memory implementation (to speed up access to data) etc within its cuda library.
ctypes/cffi/cython interoperability:
- cffi — The calling of CFFI functions is supported in nopython mode.
- ctypes — The calling of ctypes wrapper functions is supported in nopython mode…
- Cython exported functions are callable.
7. Further Reading
- https://nbviewer.jupyter.org/github/ContinuumIO/gtc2017-numba/tree/master/
- https://devblogs.nvidia.com/seven-things-numba/
- https://devblogs.nvidia.com/numba-python-cuda-acceleration/
- https://jakevdp.github.io/blog/2015/02/24/optimizing-python-with-numpy-and-numba/
- https://www.youtube.com/watch?v=1AwG0T4gaO0
8. References
- http://numba.pydata.org/numba-doc/latest/user/index.html
- https://github.com/ContinuumIO/gtc2018-numba
- http://stephanhoyer.com/2015/04/09/numba-vs-cython-how-to-choose/
Suggestions and reviews are welcome.
Thank you for reading!Signed: